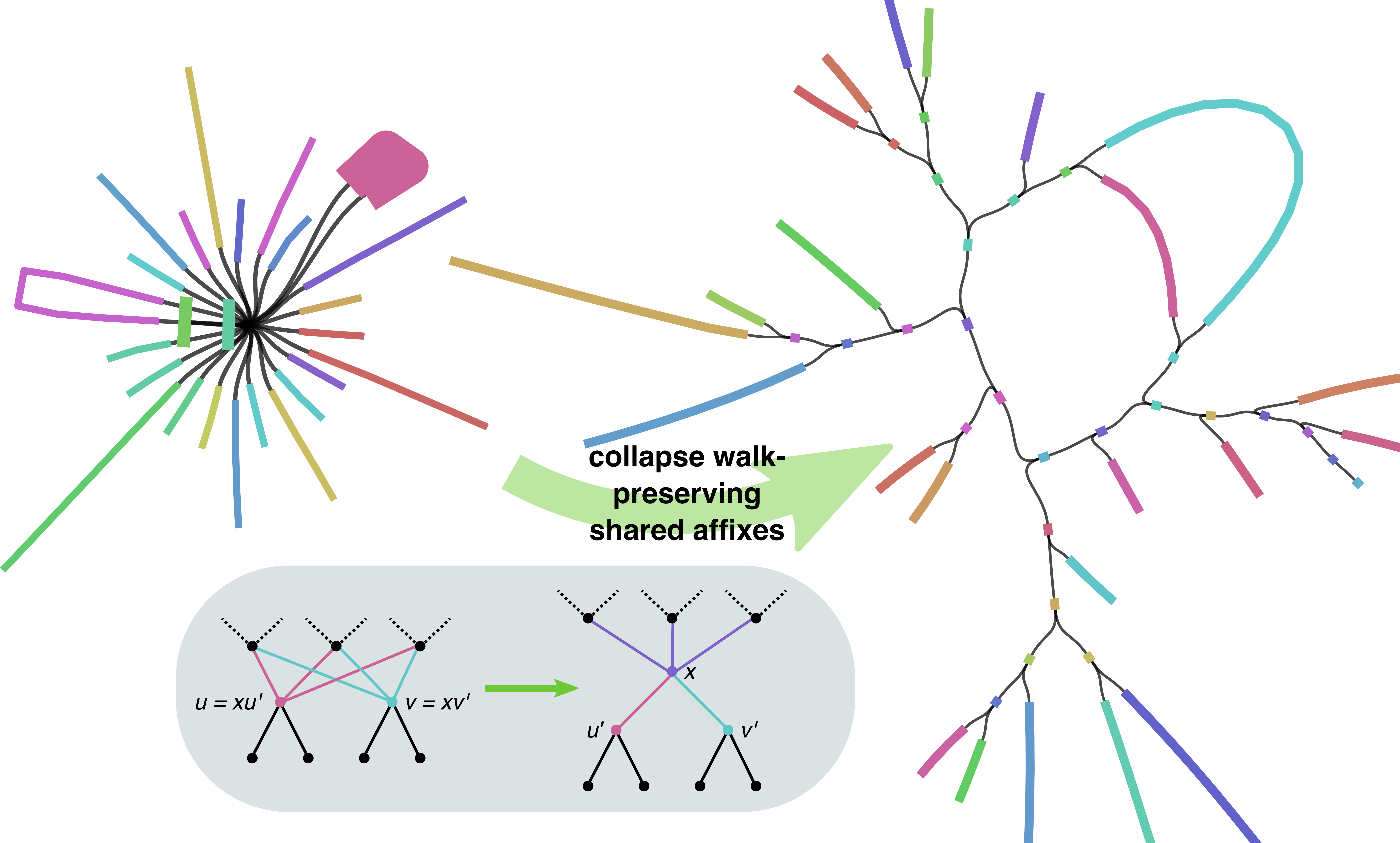
It aims to compress shared sequences that are distributed along multiple paths where one path should not have a fork (meaning we have two nodes that could be merged without any consequence on the graph information, for instance).
Publication and availability
GFAffix appears to be not published as of december 2023. A preprint is in writing (see this issue of GFAffix, but it was delayed.) Source code is available here.
Installation
Requires rust, and is available through conda.
conda create .env-gfaffix
conda activate .env-gfaffx
conda install -c conda-forge rust
conda install -c bioconda gfaffix
conda deactivateTo run GFAffix, the command is: gfaffix <input_gfa> -o <output_gfa>.
Note
The last step of pggb applies GFAffix (taken from the docs: “Finally, we apply gfaffix to remove forks where both alternatives have the same sequence.”) and minigraph-cactus applies it in it’s last step (
cactus-graphmap-join); however, if applying GFAffix on a PGGB graph returns the same graph, it is not the case for minigraph-cactus. We can expect that GFAffix is not the last step ofcactus-graphmap-join, or is ran with exclusion patterns.
GFAffix and editions
From the definition of editions I came with, I wanted to see how GFAffix impacted the resulting graph and the distance to other graphs. Without any surprise as the tool is present in both pipelines, the impact of running GFAffix is marginal.
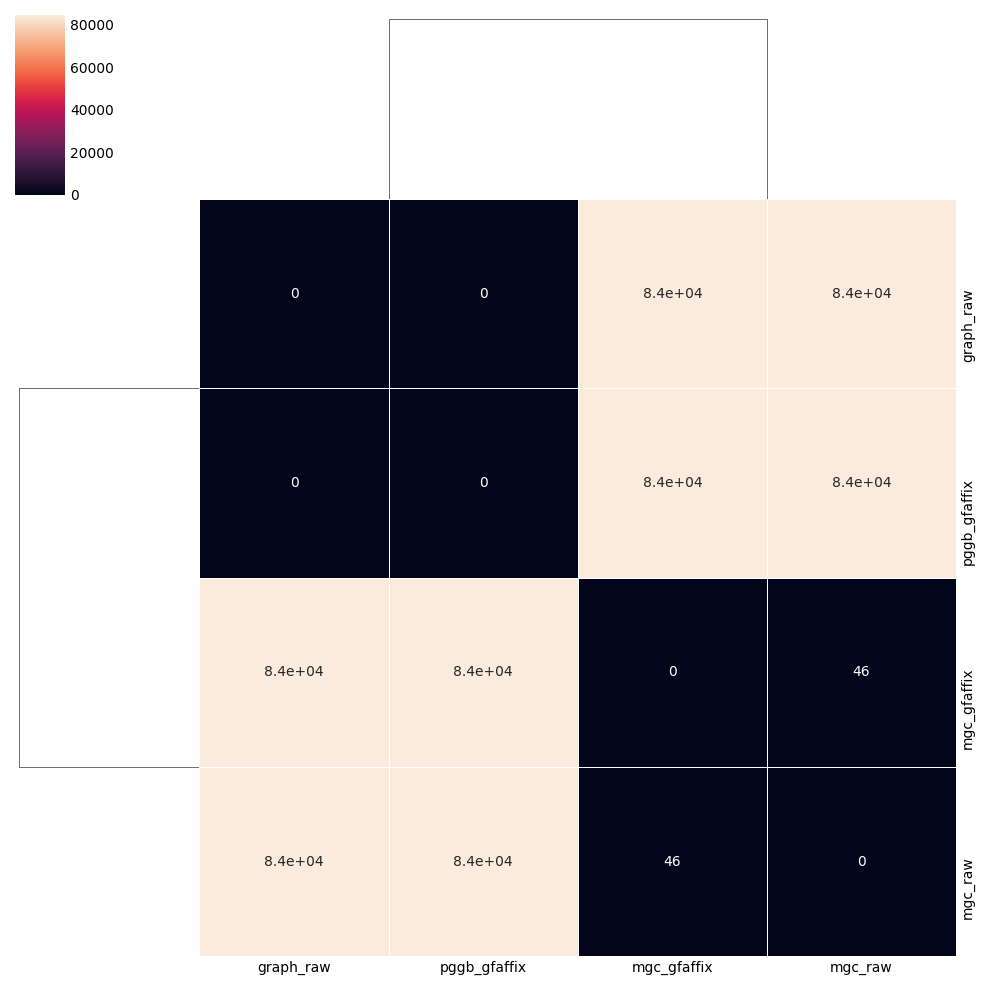
However, on graphs constructed solely using seqwish, the impact of GFAffix is not marginal: 55 editions for a graph with 820 nodes and two haplotypes